Bacteria:
| Genus | Species | Strain | Proteome Coverage |
|---|---|---|---|
| Borrelia | afzelii | Isolate B023 | 94% |
| Borrelia | burgdorferi | B31 | 96.5% |
| Borrelia | garinii | Fuji | 98.2% |
| Brucella | melitensis | Bv. 1 strain 16M | 99.5% |
| Brucella | abortus, canis, ovis, suis | various | 646 proteins |
| Burkholderia | pseudomallei | K96243 (majority), Bp576a, MSHR376b | In progress |
| Campylobacter | jejuni | NCTC11168 | 178 proteins |
| Chlamydia | muridarum | Nigg | 100% |
| Chlamydia | trachomatis | D/UW-3/CX | 100% |
| Escherichia | coli | Pan-genomic array, 98 ETEC isolates | 5,110 proteins |
| Escherichia | coli | EHEC, EPEC, EIEC, EAEC, ExPEC – specific | ~2,200 proteins |
| Franciscella | tularensis | SCHU S4 | 95% |
| Group B Streptococcus | 2603 V/R | 90 proteins | |
| Leptospira | interrogans | Serovar Copenhageni, strain Fiocruz L1-130 | 94% |
| Mycobacterium | tuberculosis | H37Rv | 96% |
| Neisseria | gonorrhoeae | various | 509 proteins |
| Neisseria | meningitidis | various (core genome) | ongoing |
| Salmonella | enterica Typhi | Ty2 | 98% |
| Shigella | boydii, dysenteriae, flexneri, sonnei | Pan-Shigella (based on sequence of ~500 isolates) | Conserved core Shigella genome (~3,300 proteins) |
| Staphylococcus | aureus (MRSA) | USA300 | 90.3% |
| Streptococcus | pneumoniae | Pan-genomic (based on sequence of ~600 isolates) | 98% (variable gene diversity) |
| Treponema | pallidum, subspies pallidum | Nichols | 99% |
Virus:
| Name | Serotype | Strain | Proteome Coverage |
|---|---|---|---|
| Chikungunya | 06-049 | 100% | |
| Dengue | Type 1 | Type1/Hawaii VR1254 | 91% |
| Dengue | Type 2 | Type2/New Guinea C | 100% |
| Dengue | Type 3 | Type3/H87-3 | 100% |
| Dengue | Type 4 | Type4/H241 | 100% |
| EBV | Varies | 195 proteins | |
| HIV | 1 | 9 subtypes | 82% |
| HIV | 2 | 2 subtypes | 96% |
| Human Adenovirus | 40 | strain Dugan | 100% |
| Human Adenovirus | 41 | Strain Tak | 100% |
| Human Coronavirus | 229E | ATCC VR-740D | 100% |
| Human Coronavirus | HKU1 | HKU1 | 100% |
| Human Coronavirus | NL63 | Amsterdam I | 100% |
| Human Coronavirus | OC43 | ATCC VR-1558 | 92.6% |
| HPV | 9 types | 83% | |
| Herpes simplex Virus | 1 | 93% | |
| Herpes simplex Virus | 2 | 84% | |
| Influenza | A | A/California/04/2009 & A/Victoria/361/2011 | ongoing |
| Influenza | B | B/Hong Kong/330/2001 & B/ Texas/06/2011 | ongoing |
| Mayaro virus | BeAn343102 | 100% | |
| Measles virus | Edmonston WT AF266288 | 100% | |
| Middle East Respiratory Syndrome Virus (MERS) | EMC/2012 | 100% | |
| Rotavirus A | Wa | 100% | |
| Respiratory Syncytial Virus (RSV) | USA/A2000/03/04/2000 | 100% | |
| Rubella virus | RA27/3 | 80% | |
| SARS-CoV | Tor2 | 20% | |
| SARS-CoV-2 | USA-WA1/2020, HKG/VM20001061/2020, ITA/INMI1/2020 | 100% | |
| Vaccinia | WR | 99% | |
| West Nile Virus | TX 2002.1 | 100% | |
| Yellow Fever Virus | Strain 17D | 100% | |
| Yellow Fever Virus | Tiled proteins | Strain 17D | ongoing |
| Zika virus | MR766 | 100% | |
| Zika virus | Tiled proteins | MR766 | 100% |
Protozoa:
| Name | Species | Strain | Proteome Coverage |
|---|---|---|---|
| Babesia | microti | LabS1 | 91% |
| Cryptosporidium | parvum, hominis, meleagridis | Iowa II, ... | 1,720 proteins |
| Onchocerca | volvulus | Cameroon | 351 proteins |
| Plasmodium | falciparum | 3D7 | 98% |
| Plasmodium | vivax | Sal-1 | 40% |
| Plasmodium | yoelii | 17XNL | ongoing |
| Plasmodium | berghei | ANKA | ongoing |
Human Proteome:
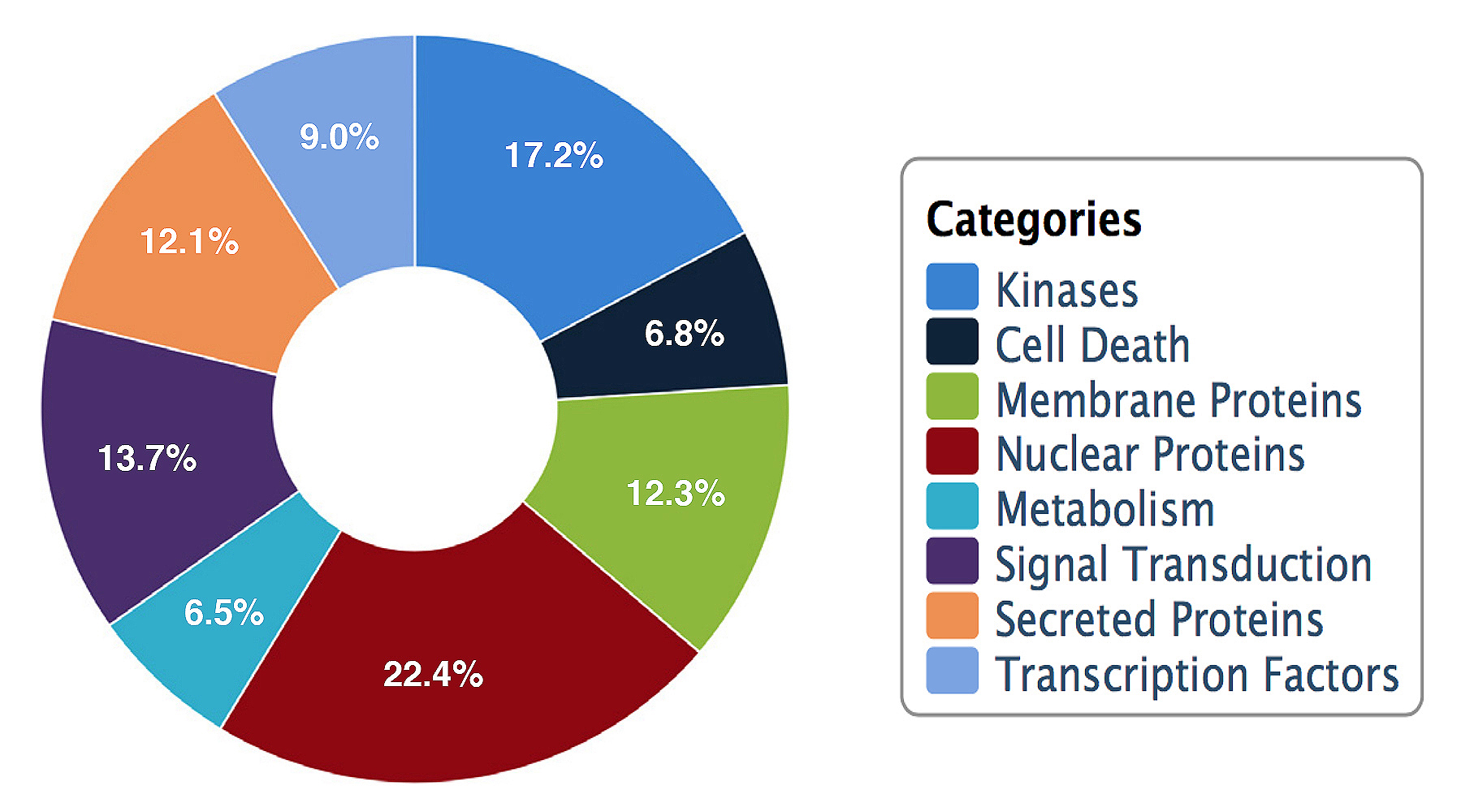
Protein Categories represented on the HuProt™ Microarray
The HuProt™ V4.0 single slide high-density format array provides greater than 21,000 proteins and protein isoforms including >81% of canonically expressed proteins as defined by the Human Protein Atlas, for rapid profiling of samples. These recombinant proteins are expressed in yeast S. cerevisiae with an N-terminal GST-RGS-His6 tag, purified, and printed in duplicate onto glass slides that are coated with an ultra-thin layer of nitrocellulose film for the non-covalent, yet irreversible, capture of active proteins to the surface. The HuProt™ arrays provide high-throughput, low sample volume screening for applications such as immune profiling for biomarker detection. In addition, the comprehensive array format can be utilized in determining protein-protein interactions for protein pathway analysis.
“New organism” and Partnership opportunities
Don’t see what you are looking for?
We are always interested to expand our current proteome collection.
Please contact us and let us know which organism you are interested in!
